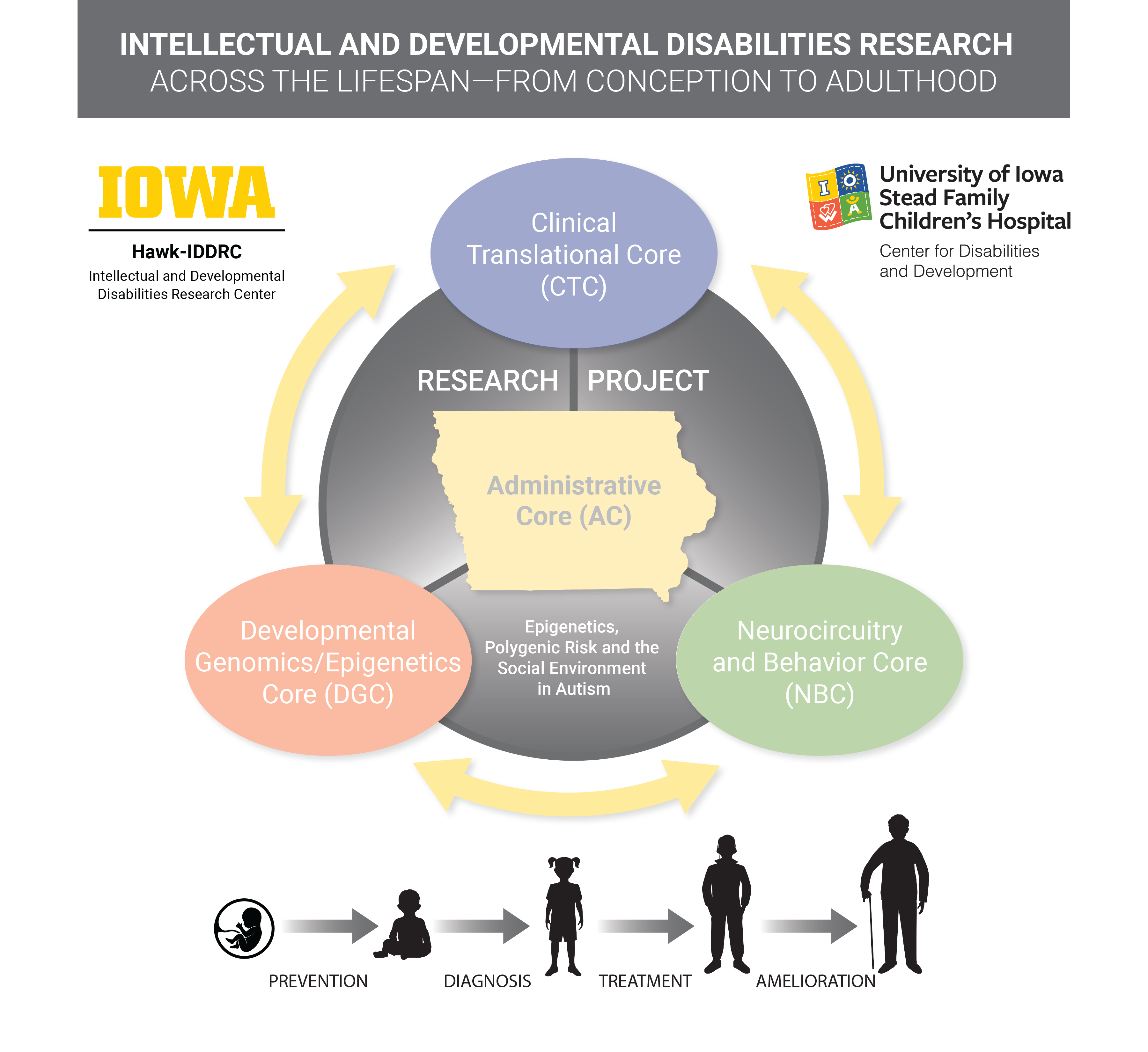
The Hawk-IDDRC Research Project will examine the interaction of genetic or environmental risks in young children with developmental disabilities, including autism, and integrate services from all four research Cores; and will integrate and capitalize upon strong existing resources in the Hawkeye State:
1) the nationally-renowned Center for Disabilities and Development;
2) the Iowa Neuroscience Institute;
3) Iowa’s University Center for Excellence in Developmental Disabilities (UCEDD), and
4) Iowa’s Leadership Education in Neurodevelopmental Disabilities (LEND) program.
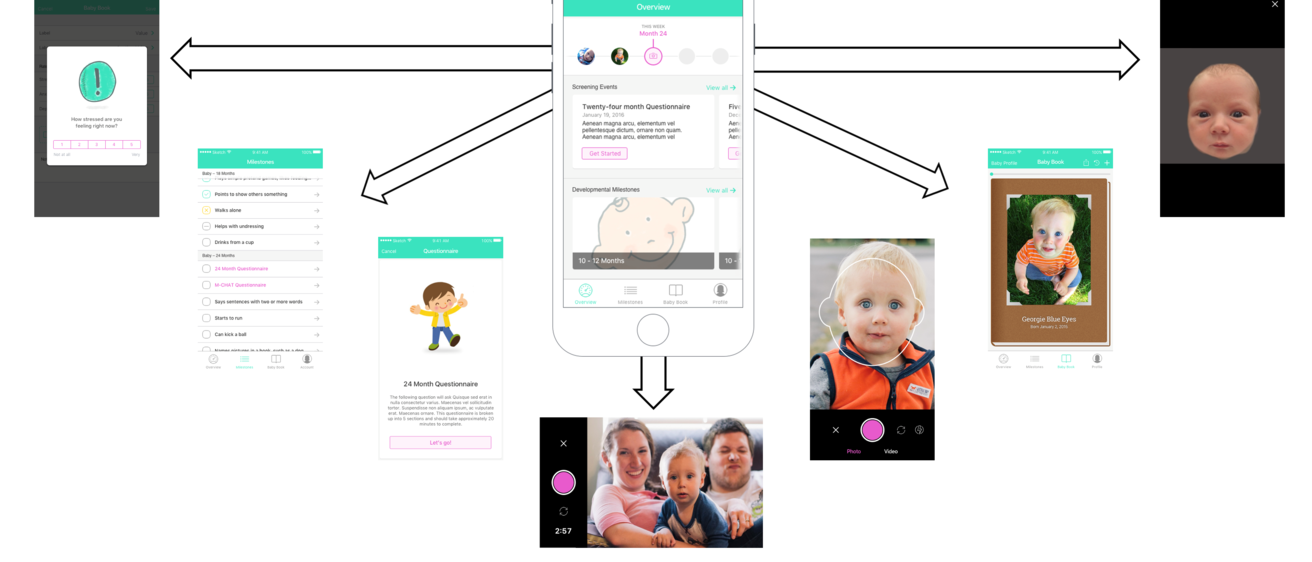
Intellectual and developmental disabilities (IDD) affect up to one in six children in the United States, with proposed causes ranging from genetic to multi-factorial. For a vast majority of children, no clear genetic or environmental etiology is identified, although evidence suggests that genetic (intrinsic) risk may interact with the early learning and social experience (extrinsic risk) to predict neurodevelopmental outcomes in later stages of life. For example, the neuropeptide oxytocin and its receptor (OXTR) are epigenetically altered by early social experience, play crucial roles in mammalian social and cognitive development, and are associated with both genetic and epigenetic risk for autism spectrum disorder (ASD). The objective of this Hawk-IDDRC Research Project is to examine epigenetic, polygenic and environmental risk factors for IDD, focusing on ASD as a prototype. We will use an innovative new smartphone application, BabySteps, to enroll and collect data on 300 mothers and their children between 2 and 3 years of age, over a 6-month period. One hundred and sixty ASD children will be enrolled, along with 80 children with non-ASD developmental delay (DD), and 80 typically developing (TD) control children, matched by race/ethnicity, sex and family socioeconomic status. We will examine changes in DNA methylation (DNAm) in 27 specific ASD-associated loci, as well as 3 OXTR loci, as a function of age and social experience, in children with and without autism, comparing DNAm from stored newborn dried blood spots to samples collected at the time of diagnosis. Polygenic risk scores for ASD will be calculated using Illumina PsychArray, and the relative polygenic and epigenetic risk assessed using resources from the Developmental Genomics/Epigenetics Core. The relationship between DNAm and social experience will be assessed using 2 complementary measures: 1) the Language Experience Analysis (LENA) based on audio recordings from a LENA device worn by the child over a 4-day period at home and in daycare; and 2) ecological momentary assessments (EMAs) of parental social and emotional availability collected using the BabySteps app. Children will be recruited through the Clinical Translational Core and Autism Center after ASD is either confirmed or excluded, based on ADOS testing and/or rigorous clinical assessment. A subset of 20 ASD and 20 TD children will participate in pilot resting state functional connectivity scanning through the Neurocircuitry and Behavior Core, to examine associations between DNAm of ASD-associated loci, including OXTR, and differences in brain connectivity. With the Administrative Core, we will share the BabySteps app with other Centers in the IDDRC Network and prospectively compare the effects of perinatal stress and social/learning experience on DNAm in children yet to be diagnosed with ASD. It is hypothesized that both polygenic factors and early social experience (via epigenetic modifications) contribute to ASD or IDD risk. Understanding how the environment interacts with genetic vulnerability will provide an unprecedented opportunity to re-assess early intervention and prevention strategies.